Computational literature-based natural product drug discovery
Researchers
Description
Natural products such as medicinal plants and extract mixtures are an important source of pharmaceuticals. In 2015, Tu Youyou was awarded the Nobel prize in medicine for her discovery of artemisinin isolated from a natural product as a treatment against malaria, saving millions of lives.
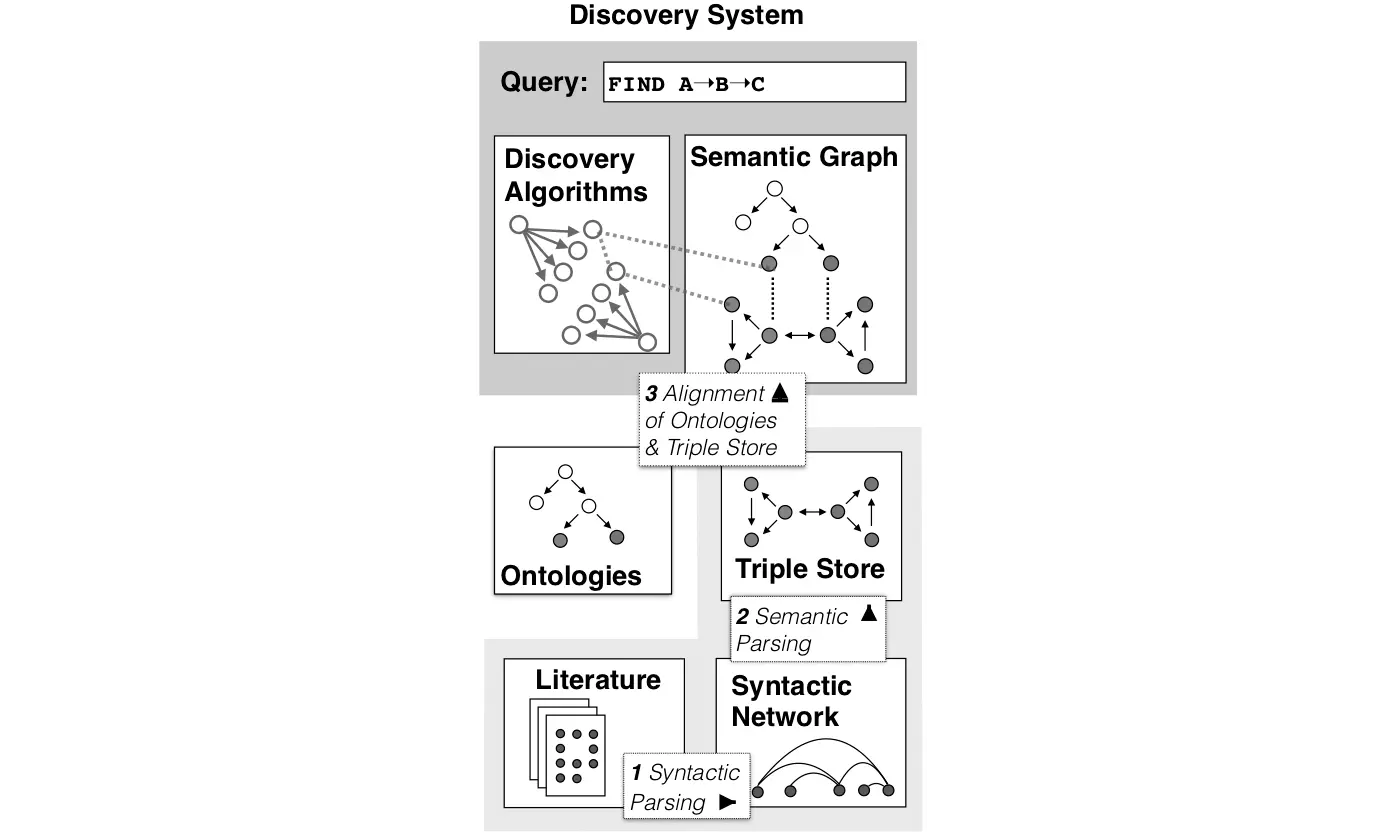
Medically relevant substances and their properties are often found through systematic, time-consuming literature analysis, nowadays referred to as Literature Based Discovery (LBD). In his pioneering work in the 1980s, Don Swanson found hidden links between unrelated pieces of knowledge in the scientific literature. He found, for example, a publication showing that fish oil (A) can reduce vascular reactivity (B), and a different publication showing that a reduction in vascular reactivity (B) can treat Raynaud’s syndrome (C). Such knowledge can be linked. From A➝B and B➝C one can deduce A➝C, i.e. consumption of fish oil could benefit patients with Raynaud’s syndrome. Swanson hypothesized this novel link, which was later confirmed through clinical trials.
Although Literature Based Discovery (LBD) has become widespread, little has been done to automate it in the field of Natural Product Drug Discovery. Today manual text mining is common practice in this field, but it is extremely laborious and time-consuming. Semantic Web technology is ideal for automating LBD. Yet, no LBD system has adopted it in an integrated approach. It builds on semantic atomic data entities, so called subject-predicate object triples. Terms in triples can be mapped to ontologies, which define a common vocabulary, machine interpretable concepts and logic based relations. The biomedical community has been very active in ontology development.
In this project researchers devise an automated LBD system for natural product drugs using Semantic Web technologies. To this end, they develop and employ state of the art natural language processing (NLP). The system will reduce discovery time by saving time in evaluating literature for drug development and is also relevant to safety and pharmacovigilance aspects
The project is an interdisciplinary collaboration between natural products research (Evelyn Wolfram, ICBT) and computational science (Manuel Gil, IAS). In addition, a PhD student and Master students from the Applied Computational Life Sciences specialisation at ZHAW also contribute to the project.