New graph classification method improves applications like drug discovery
Researchers from Winterthur and Venice developed a new method to improve the classification of graph-structured data: Topology-Aware Node Dropping Augmentation.
Graph Neural Networks (GNNs) are powerful models for classifying graphs and learning graph representations. However, like most deep learning models, GNNs struggle with overfitting, especially when datasets are limited. To improve GNNs’ robustness, data augmentation methods have been used to create synthetic data from existing graphs. But current graph augmentation methods, such as random node-dropping, tend to disrupt the essential structure of graphs, leading to potential performance degradation.
This challenge was addressed in a recent paper from the ZHAW Centre for Artificial Intelligence and Ca’ Foscari University of Venice, titled Topology-Aware Node Dropping Augmentation for Graph Classification. The paper introduces a novel augmentation technique called NDAUG (Node-Dropping Augmentation), which leverages the degree of nodes to selectively drop less important low-degree nodes while preserving essential graph structures, ensuring that GNNs maintain their performance.
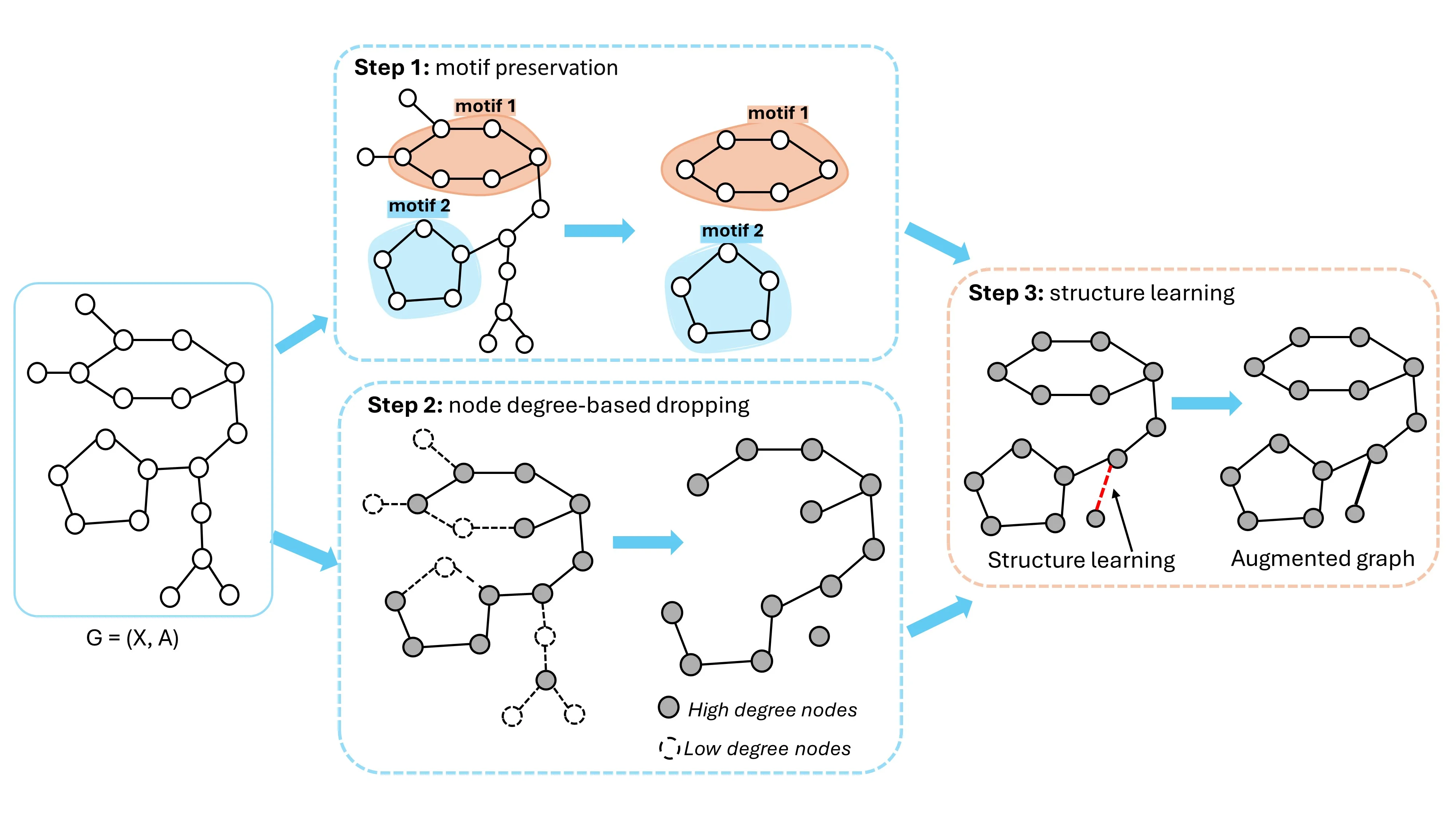
NDAUG is a three-step process:
Motif Preservation: Identifies important substructures (motifs) in the original graph, such as chemical functional groups in molecular graphs, and ensures their preservation during augmentation.
Node-Degree-Based Dropping: Drops low-degree nodes that are less crucial for the graph’s topology while maintaining high-degree nodes that represent key structural elements.
Structure Learning: Reconnects any isolated nodes resulting from the node-dropping process, using an attention-based mechanism to maintain graph connectivity.
Experiments across eight graph classification benchmarks, including datasets for molecular classification and social network analysis, demonstrate that NDAUG outperforms traditional augmentation methods like NodeDrop, DropEdge, and GraphCrop, improving performance by 2-5% on average. For instance, NDAUG improved classification accuracy on the NCI1 dataset by 4.5% and on BZR by 6.7%, compared to state-of-the-art methods.
The key innovation of NDAUG is its ability to maintain the essential topology of graphs, especially in complex networks like social interactions or molecular compounds, where the connectivity of nodes is crucial. The method's robustness is particularly evident in domains like drug discovery, where preserving the structure of molecular graphs can directly influence the accuracy of predictions.
This paper marks an important advancement in graph data augmentation, offering a more efficient way to enhance GNNs without sacrificing the integrity of graph structures. The method also paves the way for future research into augmenting graph data in ways that preserve important structural features, thus boosting the overall performance of graph-based AI systems.
The caption text for the attached diagram/figure: Fig. 1: The pipeline of the proposed NDAUG method. The initial step identifies the important structural motifs of the input graph. Step 2 removes the low-degree nodes while maintaining the key topological structures formed by high-degree nodes. The last step generates the final augmented graph to preserve the identified significant motif structures of step 1 and applies a structure learning method to retain the connectivity of the augmented graph by reconnecting any isolated nodes resulting from the node-dropping process.
Full publication: https://digitalcollection.zhaw.ch/items/1df9eaf9-4a5a-4170-90e2-601b2d8bb893
Researchers: Prof. Marcello Pelillo, Prof. Sebastiano Vascon and Dr. Waqar Ali